mRNA sequencing service
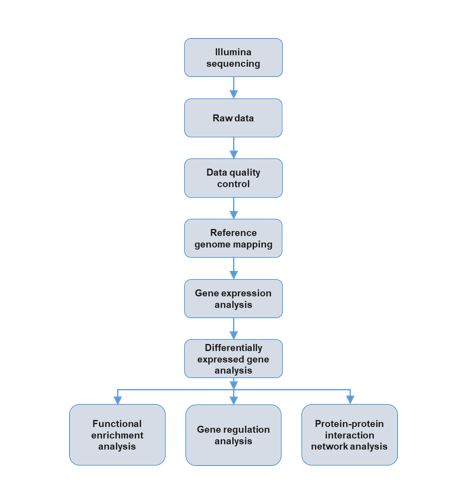
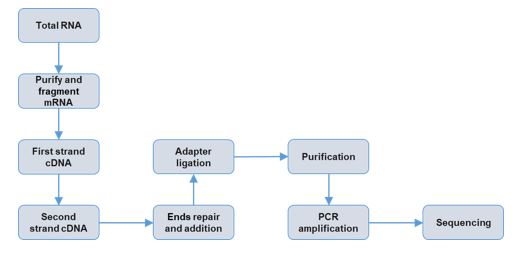
The Cancer Genomics Center of UTHealth can help you study transcriptomes of your samples via RNA-seq. The RNA you provided will be transferred to cDNA followed by ligation of adapters and then the cDNA will be sequenced by Illumina equipment. The major applications for RNA-seq include, but are not limited to, detecting differentially expressed genes, splicing, mutations at the mRNA level, gene fusions, and non-coding RNA expression.
Sample guideline
| Service | Sample type | Minimum amount | Minimum concentration | Quality |
|---|---|---|---|---|
| mRNA-seq | Total RNA | 1 µg | 20 ng/µL | RIN>7.0 |
- *RIN stands for RNA integrity number, which is evaluated using the 28S to 18S rRNA ratio.
- *Please use Qubit to measure DNA/RNA concentration.
- *Please provide Bioanalyzer results for your sample if it is available.
An Introduction to RNA-seq
Service workflow
Analysis pipeline (general purpose)