- Sequencing Service
- Bioinformatics Service
- Sample Submission
- Price
- Contact
- Resources
- About
- Careers
- A-Z
- Webmail
- Inside the University Secured Page
-
Search UTHealth Houston
Ultra-low input mRNA sequencing (Ultra-low input mRNA-seq)
This sequencing service is able to generate high-quality libraries directly from live cells (100-1,000 cells) or 1-10 ng of total RNA. It provides the excellent sensitivity needed to obtain full-length transcript information from high-quality total RNA (RIN >8) or intact cells by using oligo(dT) priming. Through even gene-body coverage and accurate representation of GC-rich transcripts, this chemistry enables reliable analysis of transcript isoforms, gene fusions, point mutations, and more.
Highlights
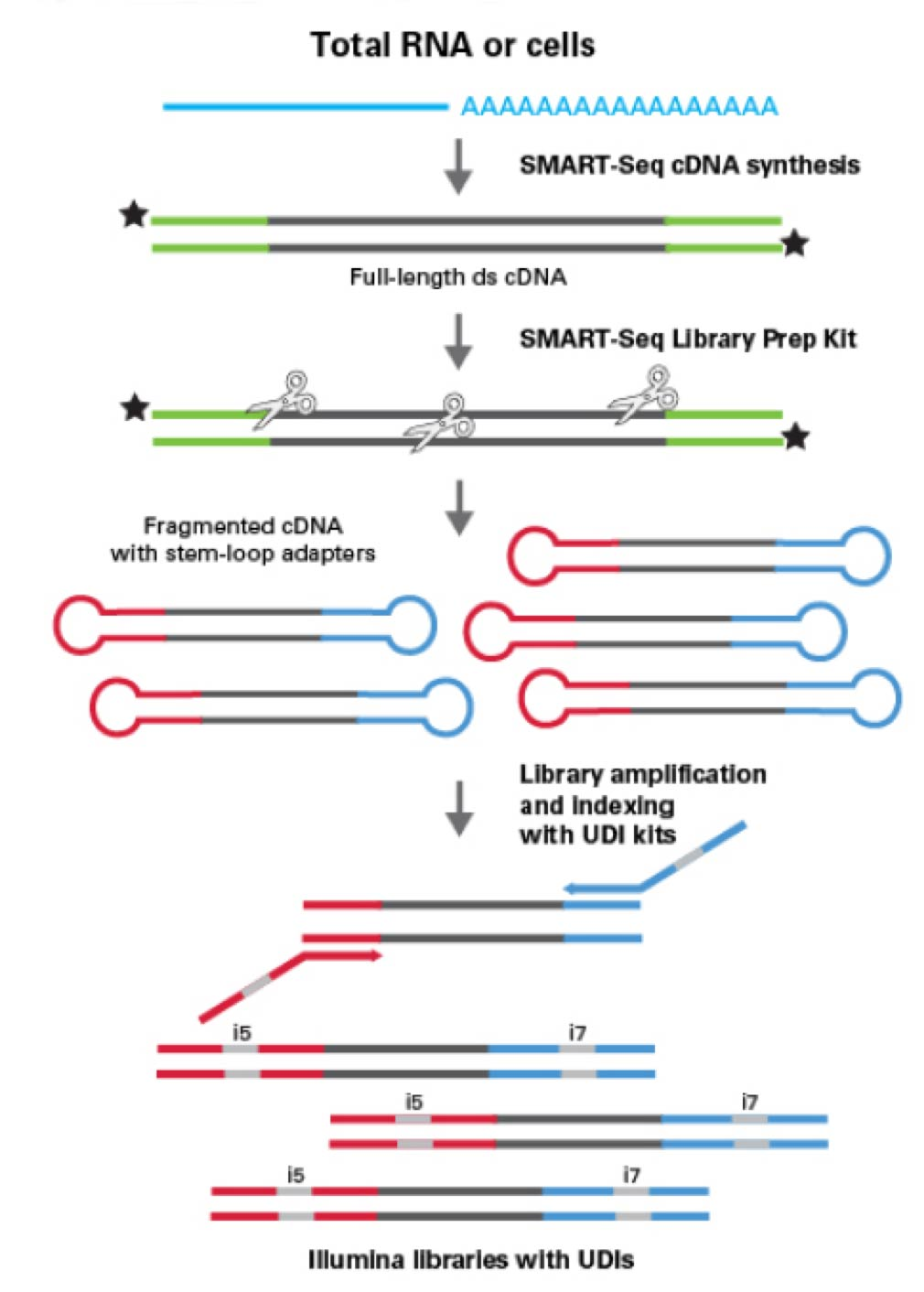
Sample submission requirements
| Service | Sample Type | Recommended Amount |
Minimum Amount |
Minimum Concentration |
Quality |
| Ultra-low input mRNA Seq | Total RNA | 10 ng | 7 ng | 1 ng/ul | Recommend: RIN>8 Required: RIN>7 |
| Live Cells | 1000-100 cells | 100 cells | Cell viability: >85% |
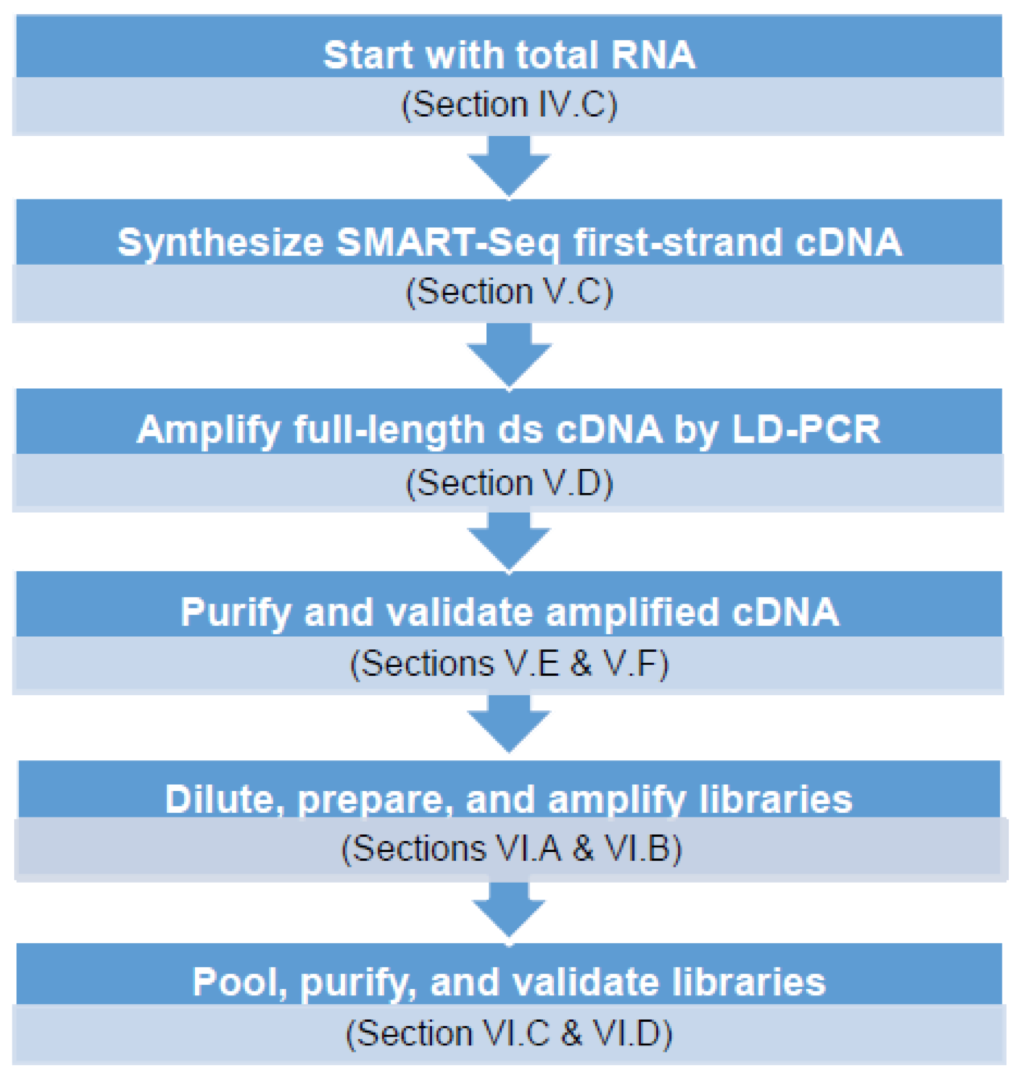
Service Workflow
Useful documents