- Sequencing Service
- Bioinformatics Service
- Sample Submission
- Price
- Contact
- Resources
- About
- Careers
- A-Z
- Webmail
- Inside the University Secured Page
-
Search UTHealth Houston
Single Cell Multiome ATAC + Gene Expression (Multiome)
Chromium Single Cell Multiome ATAC + Gene Expression provides multiomic analysis for the same single cell. Simultaneously profile gene expression and open chromatin from the same cell, across thousands of cells, with Chromium Single Cell Multiome ATAC + Gene Expression. This pipeline provides a unified view of a cell's gene expression profile and its epigenomic landscape. Increase the resolution of cell states, identify drivers of differential gene expression, and discover cells with similar transcriptional profiles but functionally different chromatin landscapes by leveraging two modalities at once.
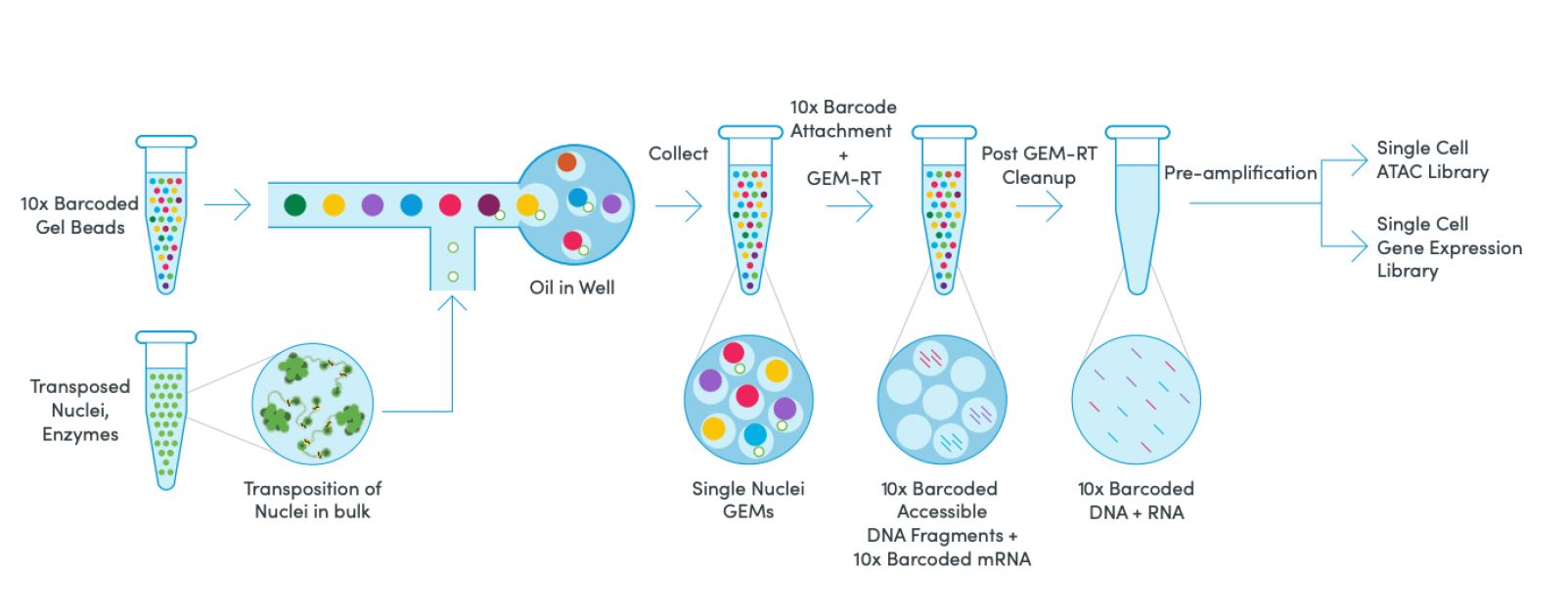
Highlights
Sample Submission requirements
| Service | Sample Type | Minimum Amount | Morphology of nuclei | Optimal concentration |
| Multime | Nuclei | 50,000 | Intact membrane | Contact us for info |
Service Workflow
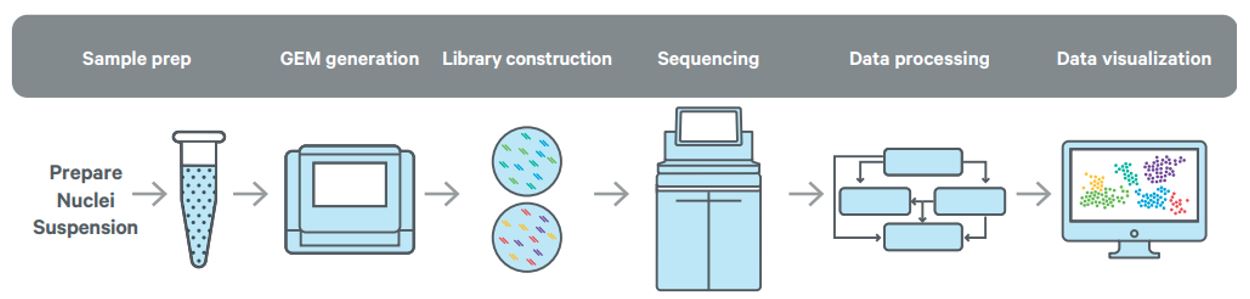
Data analysis with Cell Ranger ARC
The Single Cell Multiome ATAC + Gene Expression software suite consists of Cell Ranger ARC for the analysis and Loupe Browser for the visualization of chromatin accessibility and 3' gene expression data from the same cell produced by the 10x platform. The set of analysis pipelines in this suite perform sample demultiplexing, barcode processing, identification of open chromatin regions, and simultaneous counting of transcripts and peak accessibility in single cells. Additionally, the pipelines perform secondary analyses like dimensionality reduction, clustering, differential analysis, and feature linkage between peaks and genes that aid in data interpretation.
Useful documents
Sample preparation webinar
Recording: https://10xgenomics.zoom.us/rec/share/WBreUC7kp8YBphiFiBm3caNY4lVREJzkGK9eH-yvgtWqHV72O9fGH50xoejM8coi.nIy34oQhfX-q8tVZ