Cell Multiplexing Single Cell RNA sequencing (CellPlex-scRNA-seq)
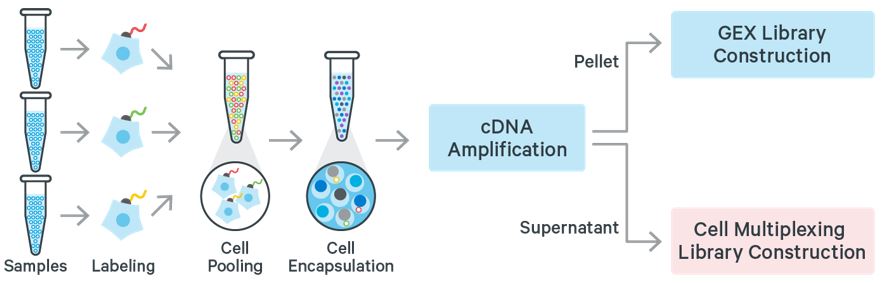
Cell Multiplexing refers to the labeling of a cell or nuclei sample with a molecular tag and subsequently mixing this sample with other labeled samples. This set of multiplexed samples can be processed together in a single GEM well. After cell encapsulation, library preparation, and sequencing, molecular tag information can be assigned to cells. Tag assignment enables identification of droplets that originally contained one (singlet) or more cells (multiplets). Cells assigned a given single tag are binned together, bioinformatically recapitulating the individual samples originally mixed together.
Highlights
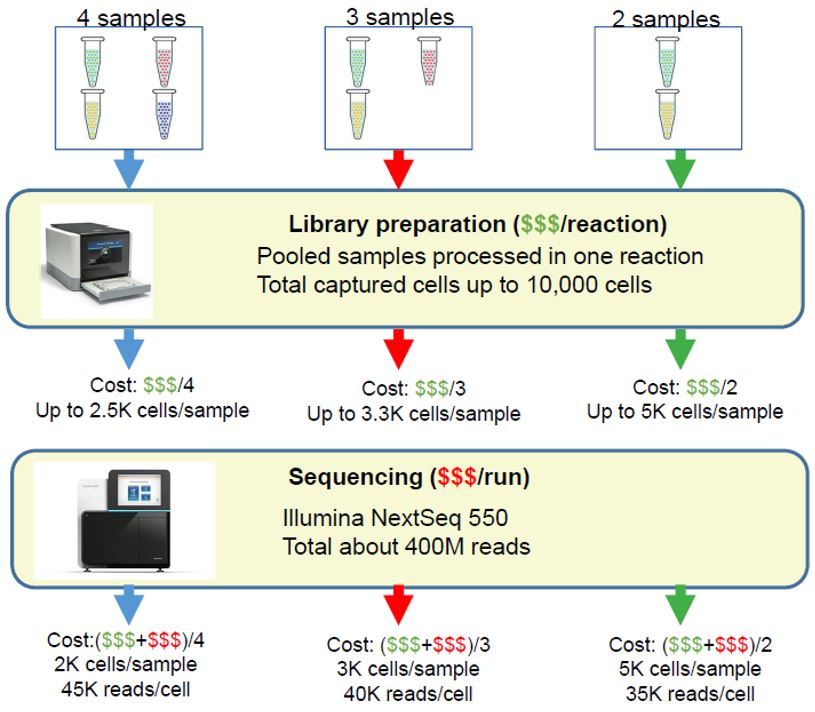
- Dramatically reduce the cost of library preparation for each sample
- Increased sample throughput in a single experiment
- Increased number of cells assayed in a single experiment
- Increased number of possible replicates in a single experiment
- Detection of multiplets and their removal prior to analysis
Sample submission requirements
| Service | Sample Type | Minimum Amount | Minimum Cell Viability | Optimal concentration |
| CellPlex-scRNA-seq | Cells | 1 million cells per sample | 85% | 700-1200 cells/ul |
- Single cells are suspended in 1xPBS with 0.04%BSA
- Samples have to be sent to core before 1pm
- The cells will be checked the quality and the core has the rights to determine if the samples are qualified for the experiment
- Please contact the core via email at [email protected] or call 713-500-7933 if your sample is isolated nuclei.
Service workflow
CellPlex-scRNA-seq (Single Cell 3' v3 kit)
Analyzing Cell Multiplexing Data with Cell Ranger
Cell Ranger 6.0 or later is required to analyze Cell Multiplexing data. Multiple samples are uniquely tagged with CMOs prior to pooling in a single GEM well, resulting in a CMO and Gene Expression (GEX) library for each GEM well. After demultiplexing the BCL files with cellranger mkfastq, run the cellranger multi pipeline on the combined FASTQ data for the CMO and GEX libraries to obtain separate per-sample output files for each CMO.