OCM scRNAseq (10x Genomics 3’ or 5’ on-chip multiplexing single-cell RNA sequencing)
The GEM-X Universal Gene Expression OCM assay (3’ or 5’) offers a scalable microfluidic platform for on-chip multiplexing of up to 8 samples—organized into two sets of up to 4 samples each. This enables the simultaneous profiling of cell surface protein expression and single-cell 3' or 5’ gene expression across 500 to 5,000 individual cells per sample.
Each cell’s transcriptome and surface protein profile are uniquely indexed using a pool of approximately 900,000 barcodes. Samples within each multiplexing set are separately partitioned with corresponding gel beads, each carrying a distinct barcode list. Nanoliter-scale Gel Beads-in-Emulsion (GEMs) are then generated in a single recovery well per set.
Within each GEM, cDNA is synthesized from polyadenylated mRNAs, and DNA is generated from cell surface proteins, with all nucleic acids from a single cell sharing the same barcode. Dual-indexed libraries are then constructed from each GEM pool, sequenced, and analyzed using 10x Barcodes to trace individual reads back to specific cells, partitions, and samples.
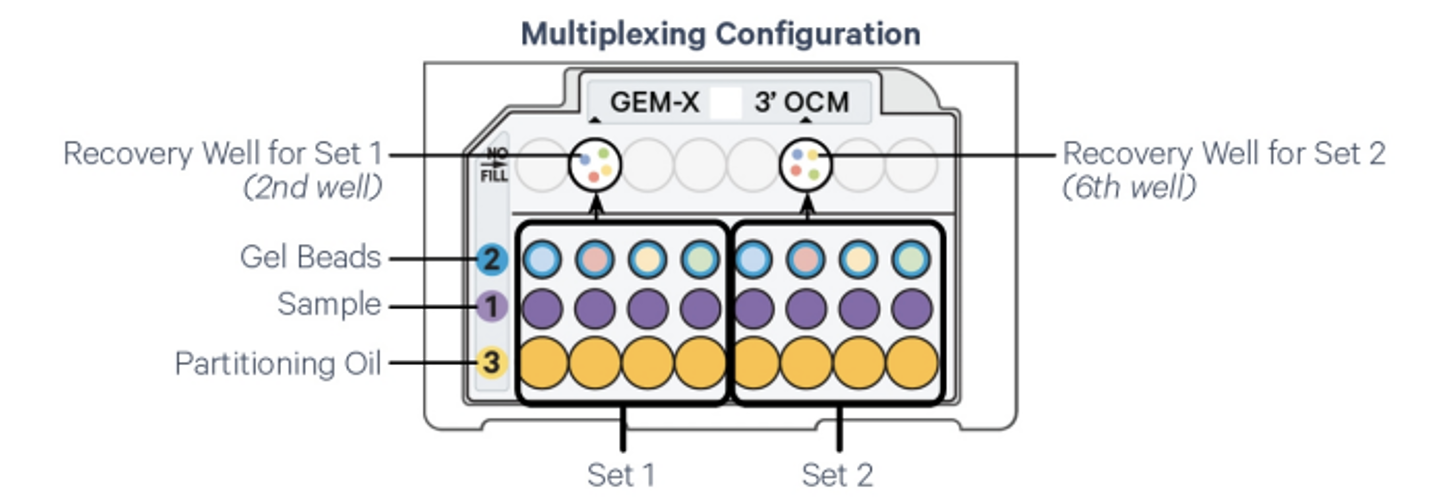
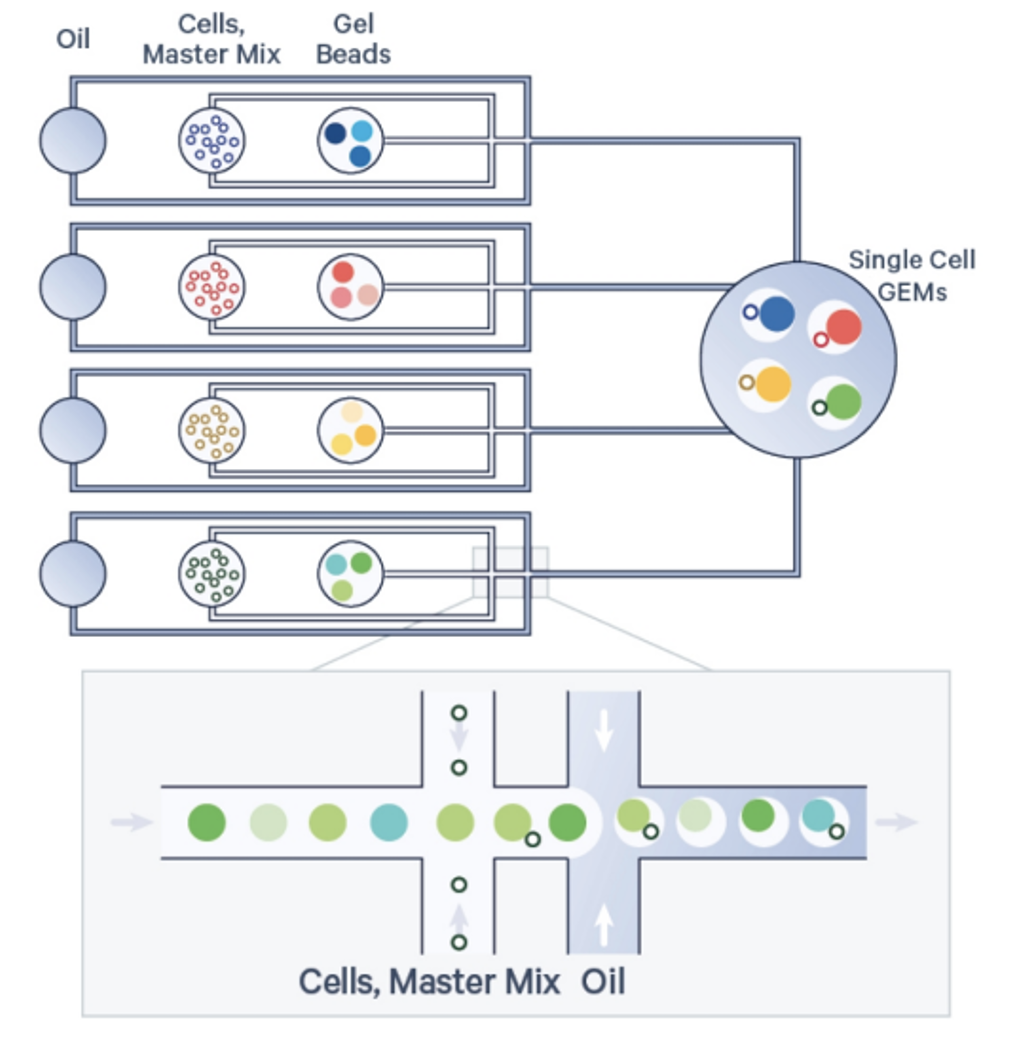
Sample Submission requirements
|
Service |
Sample Type |
Minimum Amount |
Minimum Cell viability |
Optimal concentration |
|
scRNA Immune |
Single cells |
50,000 |
85% |
700-1200 cells/ul |
- Single cells are suspended in 1xPBS with 0.04%BSA
- Samples have to be sent to the core before 1 PM
- The nuclei will be checked for quality and the core has the right to determine if the samples are qualified for the experiment
- Please check the core ([email protected] or 713-500-7933) for the detailed information.
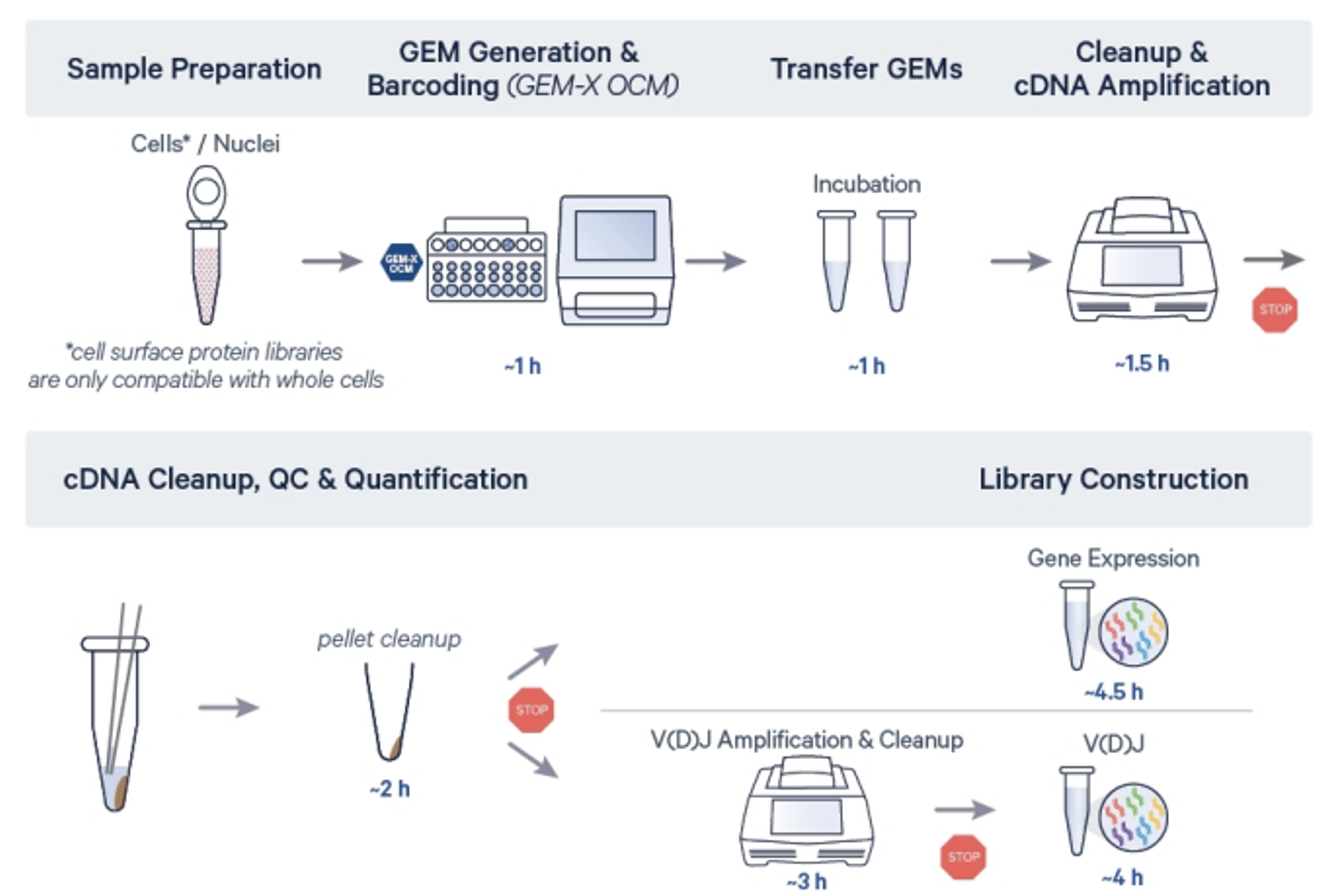
Service workflow
3’ OCM workflow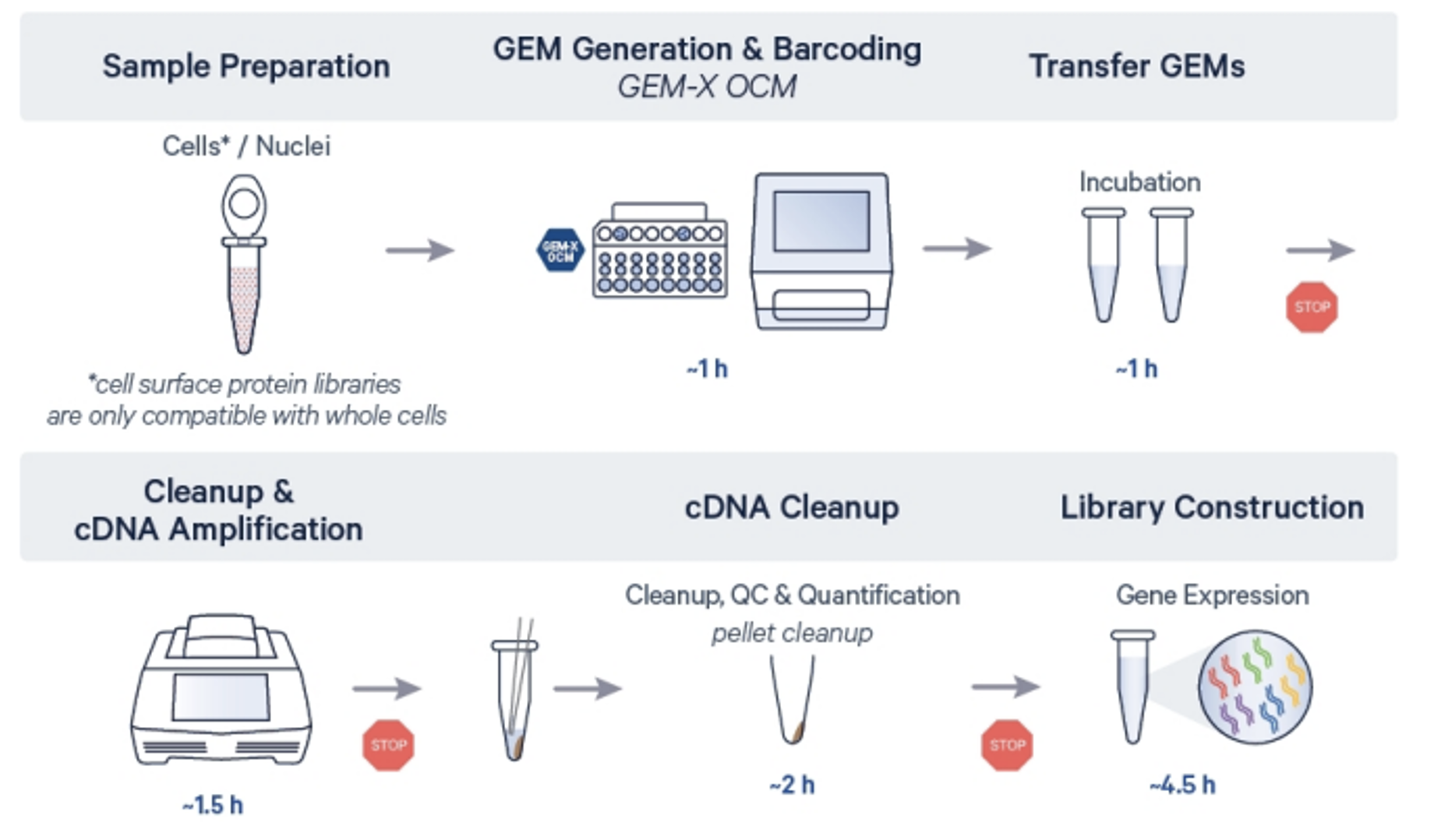
5’ OCM workflow
Data analysis
Cell Ranger is a set of analysis pipelines that processes Chromium Single Gene Expression data to align reads, generate Feature Barcode matrices, and perform clustering and gene expression analysis.
- Input: Base call (BCL) and FASTQ
- Output: BAM, MEX, CSV, HDF5, Web Summary, .cloupe/.loupe
- Operating System: Linux
Cloud Analysis is currently only available for US customers. Cloud Analysis allows users to run Cell Ranger analysis pipelines from a web browser while computation is handled in the cloud.
- Key features: scalable, highly secure, simple to set up and run
- Input: FASTQ
- Output: BAM, MEX, CSV, HDF5, Web Summary, .cloupe/.loupe.
Loupe Browser is an interactive data visualization tool that requires no prior programming knowledge.
- Input: .cloupe
- Output: Data visualization, including t-SNE and UMAP projections, custom clusters, and differentially expressed genes
- Operating System: MacOS, Windows
Useful documents