ChIP-seq service
Chromatin immunoprecipitation (ChIP), a classical experimental method for studying the interaction between protein and DNA, is widely used in related fields such as histone modification and gene expression regulated by specific transcription factors. With the development and maturity of NGS technology, ChIP sequencing (ChIP-seq), an integration of chromatin immunoprecipitation experiments with high-throughput sequencing, enables efficient and accurate screening of protein-binding sites across the genome. Furthermore, DNA binding sites associated with specialized proteins can be compared in different cell types and tissues due to the living cells originated in protein-DNA complexes.
Sample guideline
| Service | Sample type | Minimum amount | Minimum concentration | Quality |
|---|---|---|---|---|
| ChIP-seq | DNA | 10ng | 1 ng/µL | Fragmented to 100-300bp |
- *Measuring concentration with Qubit is highly recommended.
- *Please provide Bioanalyzer results for your sample if it is available.
- *Please provide the ChIP-derived DNA in nuclease free water. If any RNase is detected when running DNA chip in our Bioanalyzer 2100, the cost of cleaning up the contamination caused by RNase will be charged.
Introduction to ChIP Sequencing
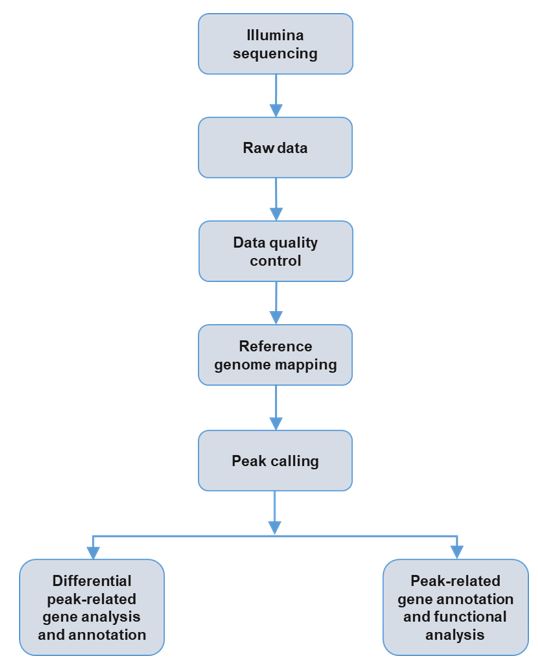
Service workflow
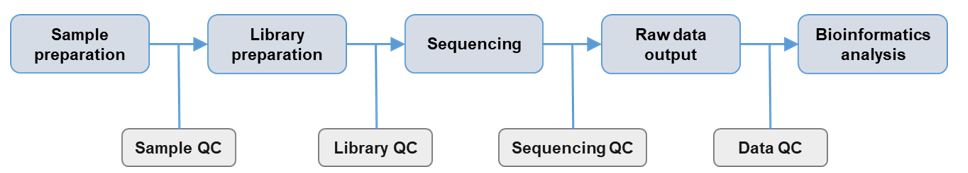
Analysis pipeline (general purpose)