Parse scRNAseq (single-cell RNAseq using fixed cells)
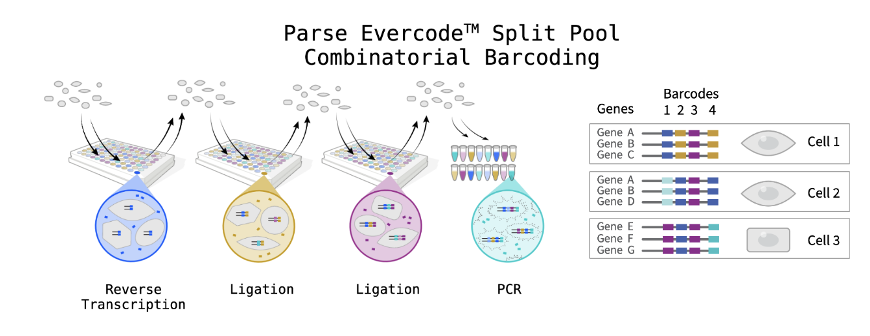
Parse scRNAseq uses Evercode™ split-pool combinatorial, which revolutionizes the way large-scale single-cell RNA sequencing experiments are conducted. The technology was based on 2018 Science paper SPLIT-seq. Cells are first fixed and permeabilized, turning them into their own reaction vessels, removing the need to capture individual cells in droplets or microwells. Then, in-situ combinatorial barcoding steps take place to create libraries ready for sequencing. The fixation protocol is able to store the samples and run them all together to reduce batch effect and give more flexibility.
Details of the final whole transcriptome sequencing library structure are below:

Highlights
- Preservation of Biological Integrity: Effectively preserves biological states and protects fragile cells by fixing fresh samples at the point of collection.
- Species Versatility: Compatible with a wide range of species, offering flexibility for various research applications.
- Cell Size Adaptability: Suitable for a broad spectrum of cell sizes, including very large cells, ensuring versatility across different cell types.
- High-Throughput Gene Expression Analysis: Enables large-scale, highly sensitive measurements of gene expression, supporting advanced research and analysis.
- Kit Options: Three kit sizes are available to accommodate varying sample sizes:
- Mini Kit: Supports up to 12 samples with a total of 10,000 cells
- WT Kit: Supports up to 48 samples with a total of 100,000 cells
- Mega Kit: Supports up to 98 samples with a total of 1,000,000 cells
Sample submission requirements
| Service | Sample Type | Minimum Amount | Morphology | Debris |
| Parse scRNAseq | Single cells or Single nuclei | 500,000 | Intact membrane | Low |
- Single cells/single nuclei are suspended in frozen media and sent on dry ice
- The cells/nuclei will be checked the quality and the core has the rights to determine if the samples are qualified for the experiment
- Please contact the core via email at [email protected] or call 713-500-7933 for more information.