Xenium In Situ Assay (spatial transcriptomics)
The Xenium Analyzer is a high-throughput instrument for single-cell spatial imaging, supported by dedicated consumables, gene panels, and software. The platform leverages advanced single-molecule RNA detection, powerful optics, and decoding technology to rapidly and accurately detect analytes across entire tissue sections with high plexity and subcellular precision. With built-in analysis capabilities, the Xenium Analyzer processes image data, localizes RNA signals, and performs secondary analysis - streamlining workflows from sample to insight. Xenium is compatible with both fresh frozen and FFPE tissue using a standardized assay that requires no optimization. Researchers can choose from a growing selection of validated gene panels, or incorporate custom gene sets, to profile up to 5,000 genes with high sensitivity.
Highlights
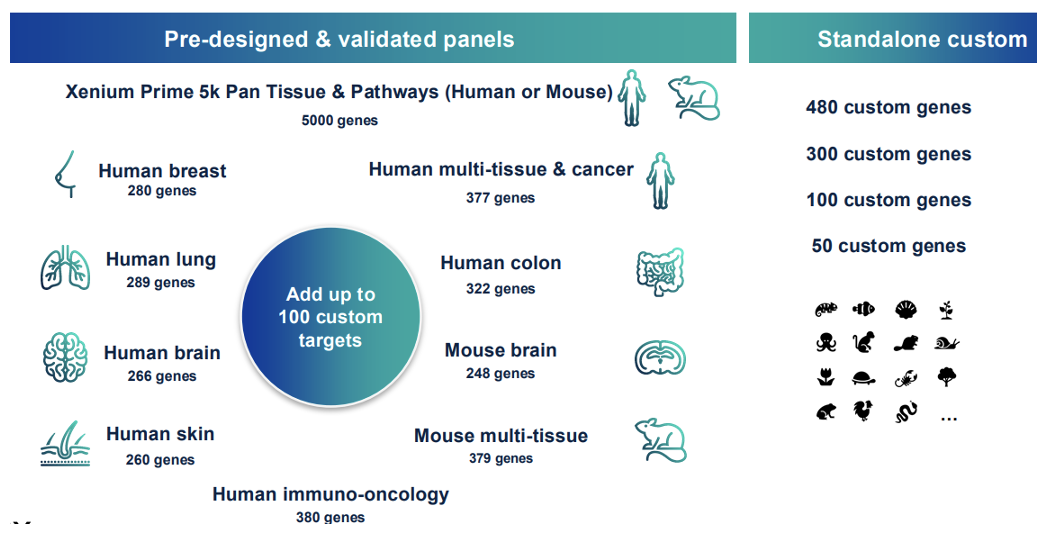
- Diverse menu of validated fit-for-purpose panels work offthe-shelf to profile up to 5,000 genes and can be customized with up to 100 custom targets; standalone custom panels of up to 480 genes can also be ordered
- Unified assay compatible with both fresh frozen and FFPE requires no optimization and is non-destructive, which allows the same section to be H&E or IF stained post-workflow
- Probe chemistry is designed to maximize specificity through dual hybridization and ligation stringency, enabling detection of expressed single nucleotide variants (SNVs) and isoforms.
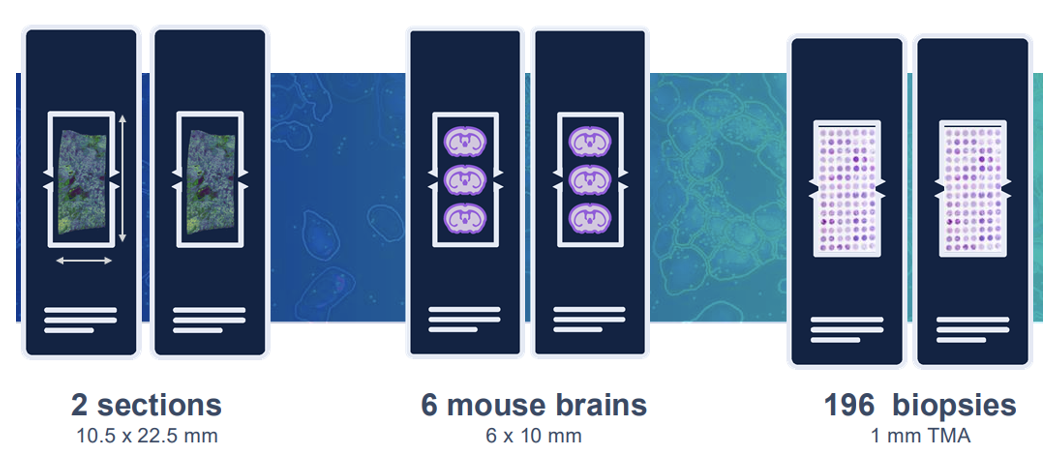
- Large 10.5 x 22.5 mm imageable area per slide
- Concurrent onboard analysis capabilities process image data, localize RNA signals, and perform secondary analysis - including multimodal cell segmentation, transcript assignment to cells, clustering, and differential expression results
Sample submission requirements
We only accept the visium slides with sections. The sample must locate inside the 10.5x 22.5 mm frame. We highly recommend to check H&E and nucleus staining using adjacent sections before the assay. Please contact the core via email at [email protected] or call 713-500-7933 for more information.
UTHealth Houston B.R.A.I.N.S. Human Biorepository and Histopathology Core or Research Histology Core Laboratory (RHCL) can help with the sample slide.
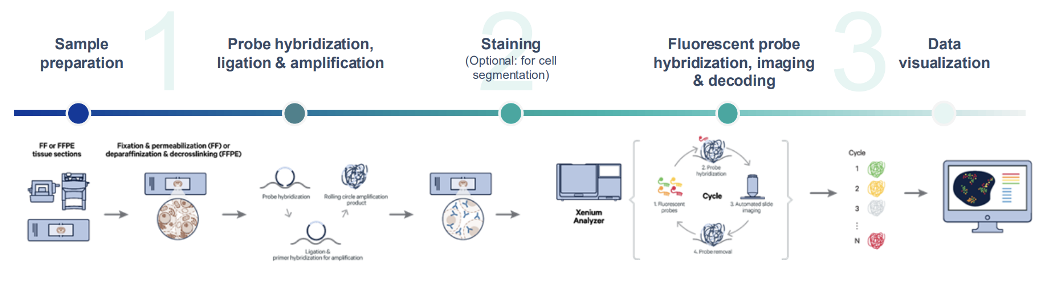
Service Workflow
Data Analysis
Onboard analysis pipeline: The Xenium Onboard Analysis (XOA) pipeline supports the analysis of Xenium In Situ Gene Expression data. The image processing and segmentation steps of the XOA pipeline depend on the assay workflow used to prepare samples. The final steps of both pipelines include RCA product image registration, decoding, deduplication, secondary analysis, and generation of Xenium Onboard Analysis output data. Quality scores (Q-Scores) are estimated using controls for calibration.
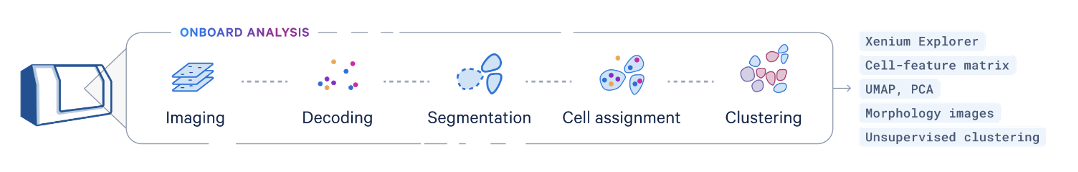
Xenium Explorer: Explore Xenium In Situ data, from tissue macrostructures to subcellular resolution, with our interactive visualization tool. Pinpoint the locations of specific transcripts, check cell segmentation, and inform downstream analysis.
Xenium Ranger: With compatibility across multiple Linux distributions, the Xenium Ranger analysis pipelines make it easy to reanalyze your Onboard Analysis data and visualize it in Xenium Explorer.