Total RNA sequencing (Total RNA-seq)
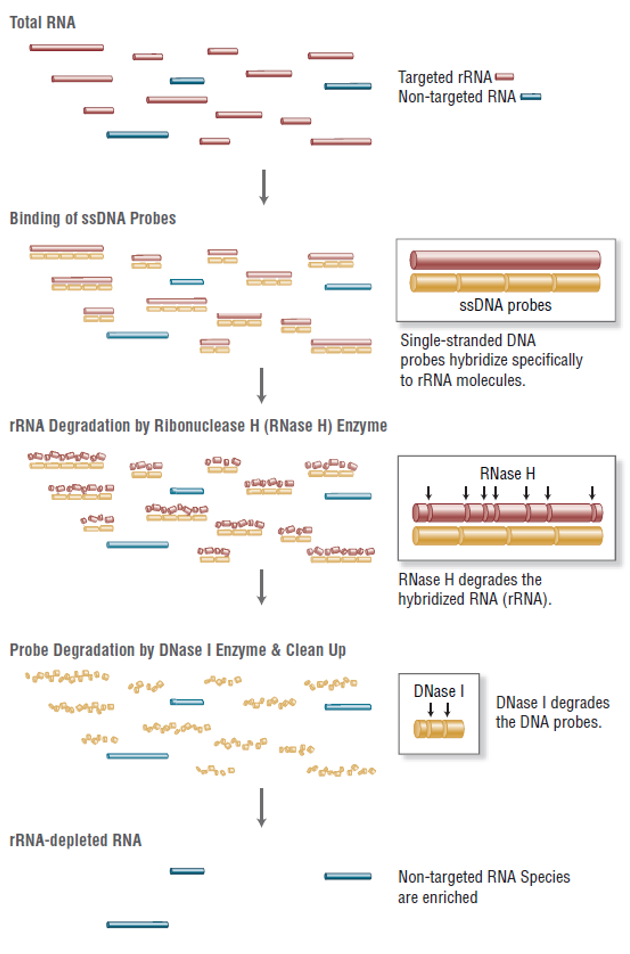
Total RNA sequencing service is able to profile both coding and long non-coding transcripts (>100bp). Long non-coding RNAs (lncRNAs) are a moderately abundant fraction of the eukaryotic transcriptome, including lincRNAs (intergenic lncRNAs), intronic, anti-sense, sense and bidirectional lncRNAs. Effects of lncRNAs show evidence on multiple cellular functions and perform as prime targets on the regulation of gene transcription, post-transcriptional modifications, and epigenetics. In total RNA-seq pipeline, mRNAs and lncRNAs were enriched by depleting rRNA from the samples as the figure below.
Sample submission requirements
| Service | Sample Type | Recommend Amount | Minimum Amount | Minimum concentration | Quality |
| Total RNA-seq | Total RNA | 1 ug | 200 ng | 40 ng/ul | Recommend: RIN>7 | Required: RIN>2.3 DV200 >66% |
- RIN stands for RNA integrity number, which is evaluated by measuring the ratio of 28S:18S
- DV200 evaluates the percentage of fragments >200 nucleotides
- Total RNA-seq may not be available for some species
- The Core has the rights to determine whether the samples are qualified for the experiment
- Please contact the core via email at [email protected] or call 713-500-7933 for more information.
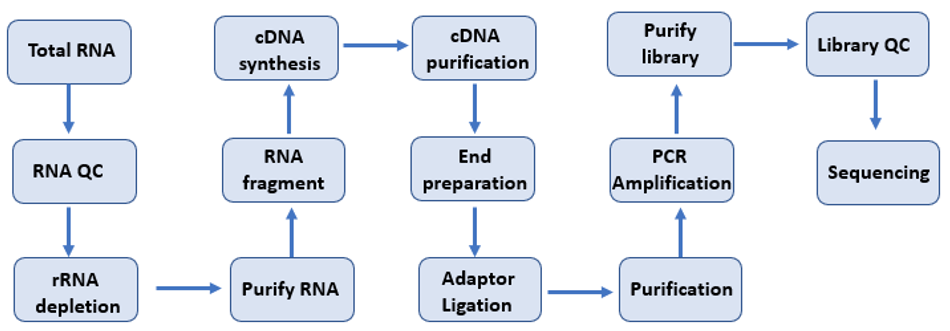
Service workflow